Research
Research vision and methodology in the TGR lab.
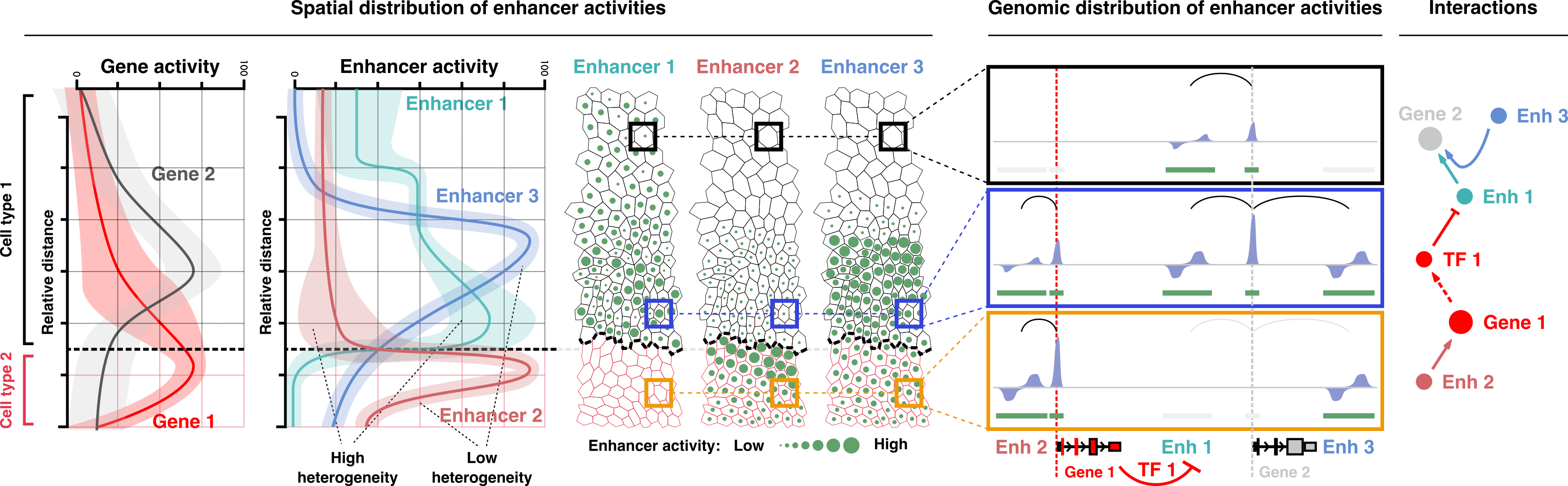
The vision of the Tissue Gene Regulation Lab is to decipher mechanisms involved in regulating gene expression and predict the activation and interaction of enhancers and promoters. However, we do not only want to resolve these activities and interactions spatially & temporarily on the genomic scale, but also on the level of spatial & temporal dynamics of entire tissues.
We understand tissue development as the sum of all gene regulatory decisions in the differentiating, proliferating, migrating and apoptosis inducing cells over time. In this framework, genetic variants cause disease by altering gene regulatory decisions (deterministic) or the possibility for such decisions (probabilistic) leading to changes in a tissue’s overall constitution such as cell composition & architecture, physiology & metabolism. To relate such manifestations of tissue dysregulation, i.e., experimental phenotypes or – more importantly – clinical symptoms to their actual cause, it is thus absolutely essential to understand the nature of the disruption elicited by the variant, including:
- The disrupted cell type(s) or cell state(s).
- The affected developmental time point along the tissue’s life trajectory.
- The spatial distribution of compromised cells in the tissue.
- The gene regulatory element(s) (i.e., enhancer(s) or promoter(s)) affected by the genetic variant and the strength with which this element’s activity is deregulated.
- The interactions with other elements and target genes of the disrupted gene regulatory element (transcriptional networks).
- The extent to what target gene(s) are misexpressed.
- The relationship between the affected target gene(s), cell type(s) and the functional, histological and morphological changes.
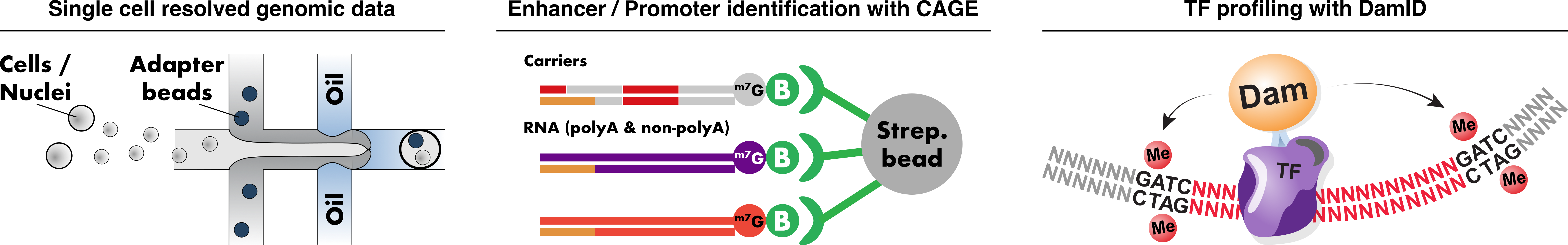
By combining our experimental and computational experience we embark on solving the first 6 points, while collaborating with other research groups and clinicians to make number 7 possible, too. For this we rely mainly on 3 high-throughput methods:
- Single cell technologies for capturing gene expression, chromatin accessibility and other modalities.
- DNA adenine methytransferase identification (DamID) to profile chromatin bound proteins in vivo and in vitro.
- Cap analysis of gene expression (CAGE) based on cap-trapping of 5’-m7G-capped RNA species – namely messenger and enhancer RNAs – to discern activated enhancers and promoters.
In the future, we will also make use of spatial transcriptomics and high-resolution microscropy.